

Our Research
We use a multidisciplinary approach to study viral protein synthesis.
We combine classical biochemistry with single-molecule fluorescence microscopy, time-resolved cryo-EM, X-ray crystallography and biophysical techniques.
When RNA viruses infect cells, they frequently make proteins that differ from the genetically encoded sequences. These highly-regulated ‘recoding’ events are vitally important to viral gene expression, and if disrupted many viruses (e.g. SARS-CoV-2, HIV-1) fail to complete their replication cycles.
A better understanding may therefore present unique opportunities for therapeutic intervention.

Fig. 1 - Conceptual overview of viral recoding
All recoding events represent a perturbation of normal elongation, and many involve a pause or stall at a “stimulatory element” before the ribosome resumes normal elongation (Figure 1A). In -1 programmed ribosomal frameshifting (PRF), a proportion of elongating ribosomes shift one nucleotide into the -1 frame, changing the amino acid sequence from that point forward (Figure 1B). During stop codon readthrough, ribosomes fail to recruit release factors at a stop codon, instead extending the protein C-terminally (Figure 1C). In StopGo translation, ribosomes release the upstream peptide and continue translation without re-initiating (Figure 1D). These events are finely-tuned to produce viral proteins in optimal ratios for efficient assembly, and they are regulated by a complex interplay between the elongating ribosome, cis-acting elements in the mRNA or nascent peptide, and trans-acting protein factors.

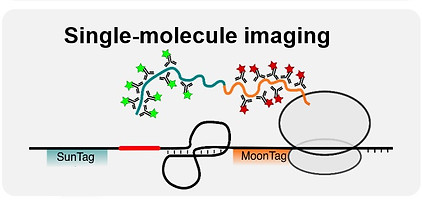
Fig. 2 - Techniques to investigate recoding kinetics
Our laboratory uses a multidisciplinary approach to better understand recoding, focussing on PRF. We combine classical biochemistry with single-molecule fluorescence microscopy to observe PRF kinetics in real-time both in vitro and in live cells (Figure 2). To reveal mechanistic details, we apply time-resolved cryo-EM, X-ray crystallography, SAXS and biophysical techniques (Figure 3) to study translating ribosomes, viral proteins, structured RNA elements and RNA-protein interactions at the atomic level. We are also interested in how cellular proteins act to inhibit recoding events as part of the interferon-stimulated response to viral infection.

Fig. 3 - Structural and biophysical approaches to study recoding
