

Our Structures
To be deposited • cryo-EM
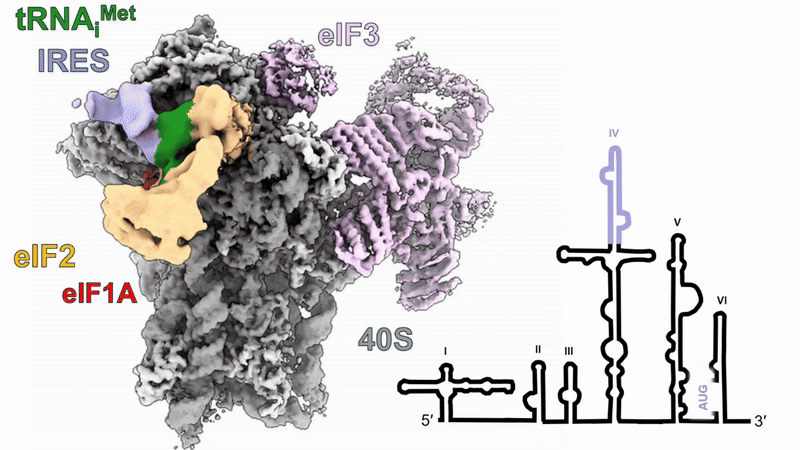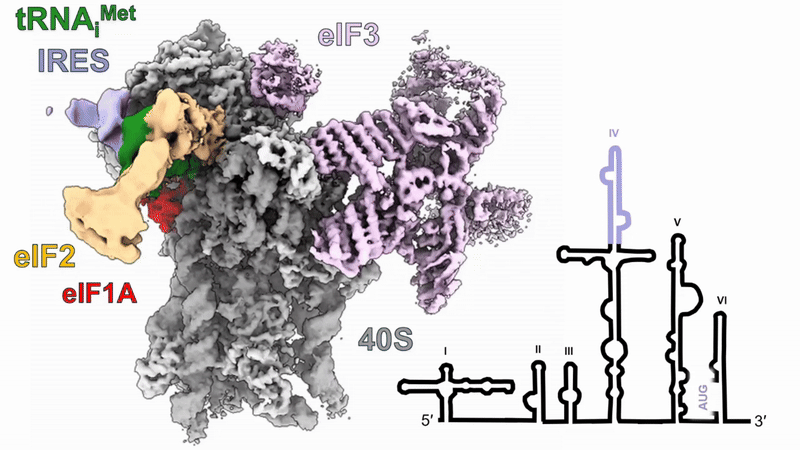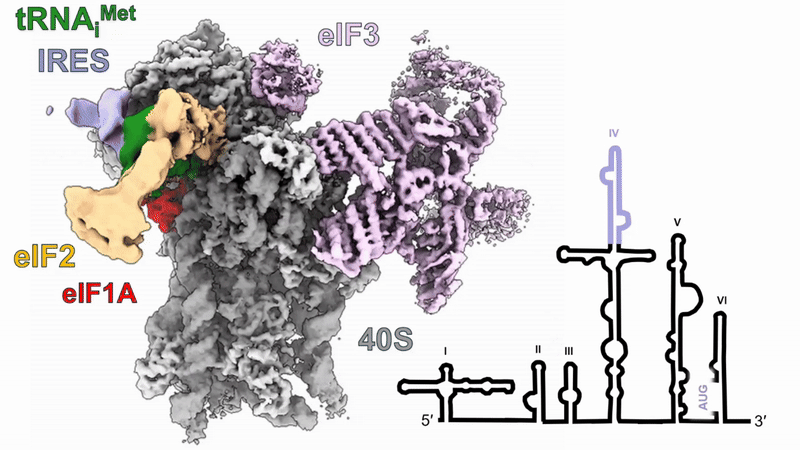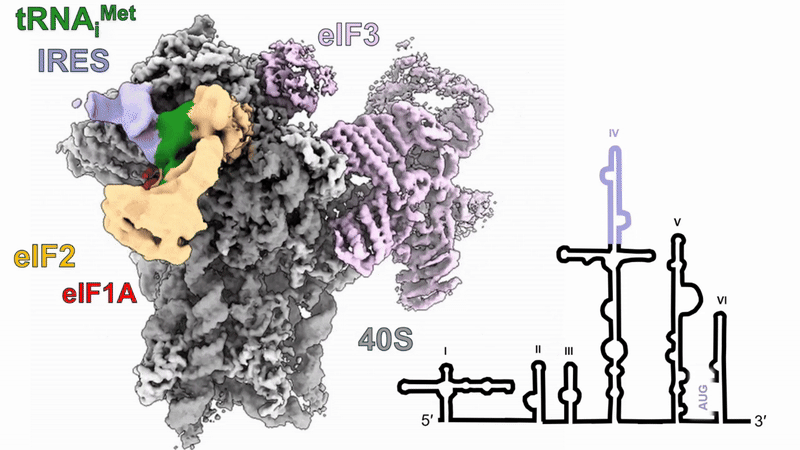
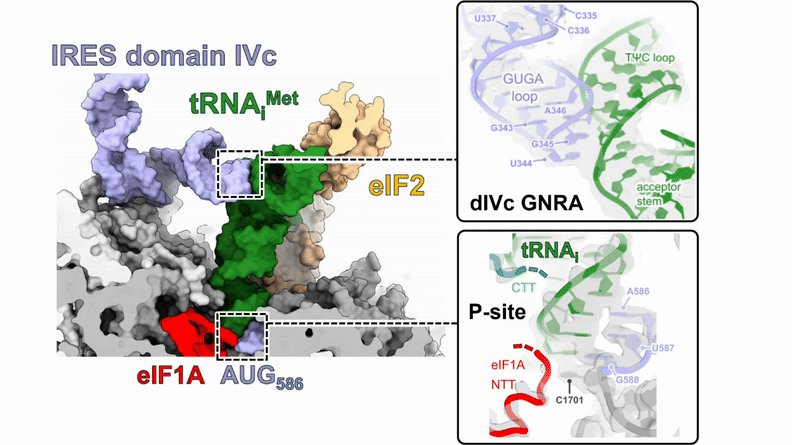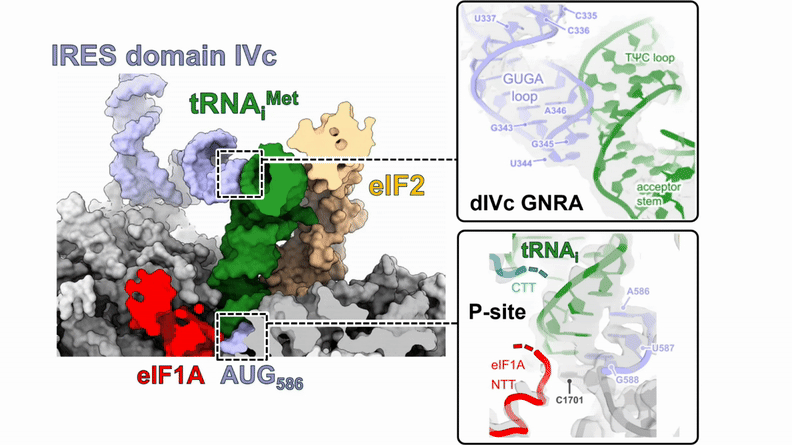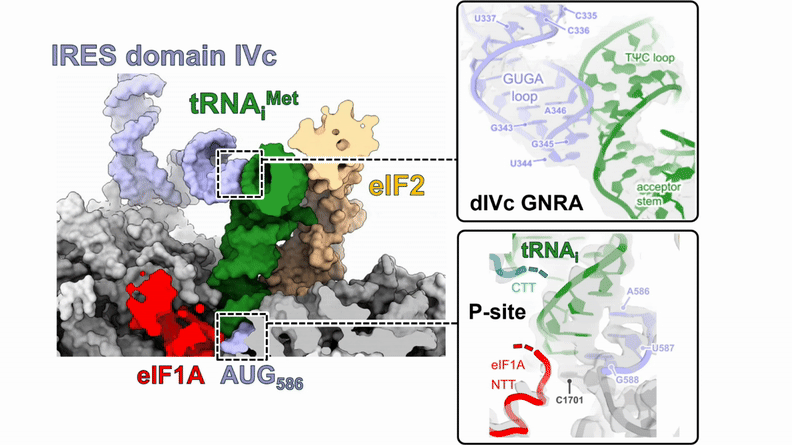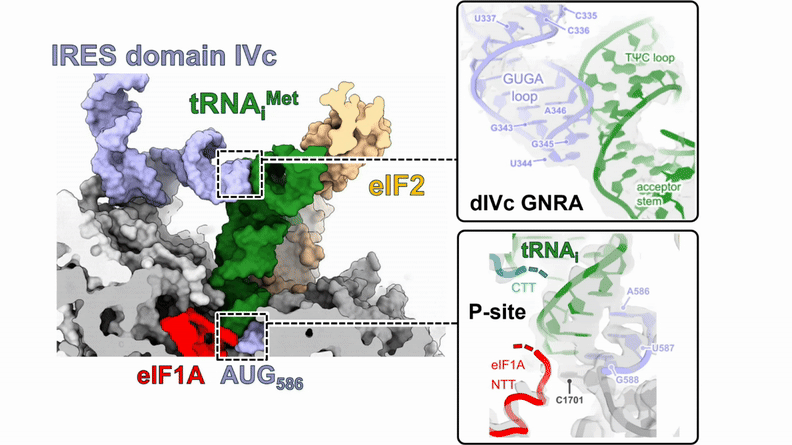
Human 48S translation initiation complexes on the poliovirus Type 1 IRES
Our structures of reconstituted human 48S complexes on the Type 1 poliovirus IRES (AUG586) reveal how IRES domain IVc contacts ribosomal proteins uS19 and uS13, whilst a conserved GNRA tetraloop engages with the acceptor arm of the initiator tRNA during start-codon recognition. Disruption of these contacts causes severe defects in IRES-driven translation and virus replication. Read more in our preprint.
9RVP • X-ray crystallography
TMEV 2A protein bound to frameshift-stimulating
RNA pseudoknot
Crystal structure of the RNA-protein complex that activates high-efficiency programmed -1 ribosomal frameshifting (PRF) in cardioviruses. The viral 2A protein binds to the RNA in a highly unusual pseudoknot conformation. Stem 1 is only three G-C base-pairs, but its formation is stabilised by insertion of the "arginine loop" residues (spheres) into the major groove, where they recognise guanine bases. See the video below for a guided tour.

8RDW • cryo-EM
P. urativorans 70S ribosome in complex with Balon and EF-Tu(GDP)
EF-Tu(GDP) was observed in ~44% of Balon-containing ribosomes. Balon uses a distinct surface on its C-terminal domain to bind to Domain III of EF-Tu. In this orientation, EF-Tu Domain III binds to the tip of the GTPase-activating sarcin-ricin loop (SRL). The closed GTP-bound conformation of EF-Tu is not compatible with Balon binding, suggesting a mechanism distinct from the usual functions of EF-Tu.


8RDV • cryo-EM
P. urativorans 70S ribosome in complex with Balon, mRNA and P-site tRNA
Balon lacks the 'NIKS' or 'loop A' motifs that allow aERF1 family proteins to bind mRNA in the A-site. Instead, it remains ~10 Å away from the mRNA channel and can therefore bind to any actively translating ribosomes. In this state, the 3' -CCA end of the P-site peptidyl-tRNA interacts with the Balon lasso loop, and the N-terminus of bL27 is sequestered by the Balon "bL27 trap", preserving the PTC in an inert state and preventing premature peptide hydrolysis.
8RD8 • cryo-EM
P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA
Balon binds in the A-site of idle unrotated ribosomes, forming contacts between the HP motif and decoding center residues A1492 (16S h44) and A1913 (23S H69). This binding mode is compatible with the simultaneous binding of RaiA to the 30S P-site. Balon extends into the PTC, but lacks the GGQ motif that triggers peptide release.


7NWT • cryo-EM
EMCV 2A protein in complex with initiated 70S ribosomes
The cryo-EM structure shows how EMCV 2A binds directly to the small ribosome subunit via direct interactions between the 2A "arginine loop", protein uS12 and 16S rRNA. Binding at this site would clash with translational GTPases, suggesting a potential explanation for the observed translational inhibition in the presence of 2A.
7BNY • X-ray crystallography
EMCV 2A protein
The crystal structure of 2A protein from encephalomyocarditis virus (EMCV) revealed a new RNA-binding fold. Interestingly, the asymmetric unit of these crystals contained four molecules with a very unusual arrangement - intermolecular disulfide bonds had formed between surface-exposed cysteine residues (yellow spheres). The "arginine loop" residues that are essential for RNA binding and frameshifting are also highlighted as spheres.
7NBV • X-ray crystallography
TMEV 2A protein
This crystal structure of 2A from related cardiovirus Theiler's murine encephalomyelitis virus (TMEV) showed that it adopts a similar 'beta-shell' fold to the EMCV orthologue, despite sharing < 20 % sequence identity. The "arginine loop" (shown as spheres) is the most conserved feature, and mechanisms of RNA recognition are likely similar.
